BioUnfold #6 — When Innovation Is Out of Sync
In BioUnfold #3, I wrote that the real leverage of computation in drug discovery comes from choosing where to build depth.
But even when teams choose the right area, progress can still feel uneven.
A common cause is simple:
Experiment and computation are often moving at different tempos.
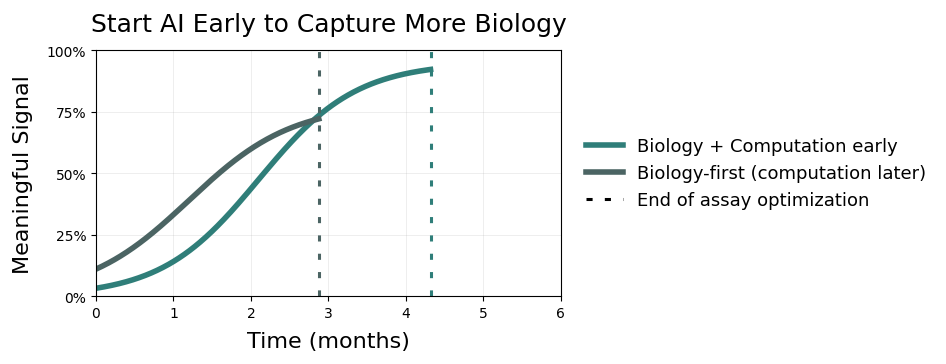
The experimental work is refining what the biological signal looks like, while the computational work is trying to understand how that signal behaves across conditions. Both are working toward the same objective — just from different angles.
The solution is not more alignment meetings. It is accepting that bringing computation in early costs time now, but that is the time that expands the biology and lets the program move faster later.
Not a Pipeline — a Rhythm
In experiment-driven discovery, computation progresses through three modes:
-
Exploration — What exactly are we measuring?
Small experiments, many hypotheses, signal and noise still entangled. -
Repeatable Interpretation — We know what matters; we apply it consistently.
Same workflow each time, QC checks, results stable but still interpreted by humans. -
Automation — We have seen this enough times that it is now just execution.
The logic is clear, inputs are stable, failure modes are known.
The work is not to push everything toward automation; it is to place each step in the mode that serves it. Automation optimizes for speed and consistency — which is only useful once we know what we want to preserve. Bringing data science in earlier often expands the number of hypotheses the team explores, which slows things down in the short term. That is not inefficiency — that is discovery.
Most real programs use these modes in parallel rather than sequentially.
Example: Immunofluorescent Screening
In immunofluorescent screens I have worked on, preprocessing and normalization could eventually be automated end-to-end.
However, the QC validation — the before/after plots, plate drift checks, and feature distribution sanity checks — remained manual. The workflow was structured and repeatable, but still reviewed by a scientist.
Why?
Because even when the math is stable, the meaning of the signal still depends on biology:
cell line adaptation, staining consistency, handling differences.
During screen optimization, the analysis is almost entirely exploratory — we are learning what signal looks like.
It only becomes repeatable during hit validation, when a pattern holds across conditions.
And once that pattern is shown to be stable, automation is useful — not because it is more “advanced,” but because it gets results the next day.
Automation is not the goal.
Automation is what you do once the thinking is done.
Where Teams Go Wrong
Misalignment happens when automation is introduced before repeatable interpretation exists.
In that case, software freezes uncertainty into the workflow — and teams end up spending more time fighting the tool than learning from the experiment.
The opposite problem also happens:
Teams stay in exploration mode far too long, running every analysis as if it were new.
This slows learning and makes collaboration fragile.
The art is in knowing when to shift the cadence.
A Simple Way to Find the Right Tempo
Take your current program and ask:
| Question | If the answer is yes… | You are likely in… |
|---|---|---|
| Are we still figuring out what “signal” means? | Run fewer models, more controlled experiments | Exploration |
| Do we mostly repeat the same analysis every cycle? | Standardize workflows and QC, don’t automate yet | Repeatable Interpretation |
| Do we already trust the outcome logic? | Automate to free time, not to add capability | Automation |
Most programs sit in more than one stage at once.
That is healthy.
The goal is not to push everything forward.
The goal is to put each step in the right place.