BioUnfold #13 — The Three Layers of Biologics
How Extracellular, Intracellular, and Genomic Mechanisms Shape Therapeutic Design
Small molecules have been the centre of this series so far. They are where most of my experience lies, and where AI has been most publicly discussed. But biologics have been redefining the therapeutic landscape for two decades, and they pose a different, often more structured, design space. Their constraints are not the same as chemistry’s. Their opportunities are not the same either.
If small molecules are engineered artefacts shaped by human creativity, biologics are biological molecules shaped by evolution and repurposed by design.
This post steps into that world.
What We Mean by “Biologics”
Biologics are therapeutics produced by cells as factories, not chemical synthesis. The category spans proteins, nucleic acids, viral vectors, and engineered cells. Yet the design space becomes clearer when grouped by where they act.
Three layers capture the landscape.
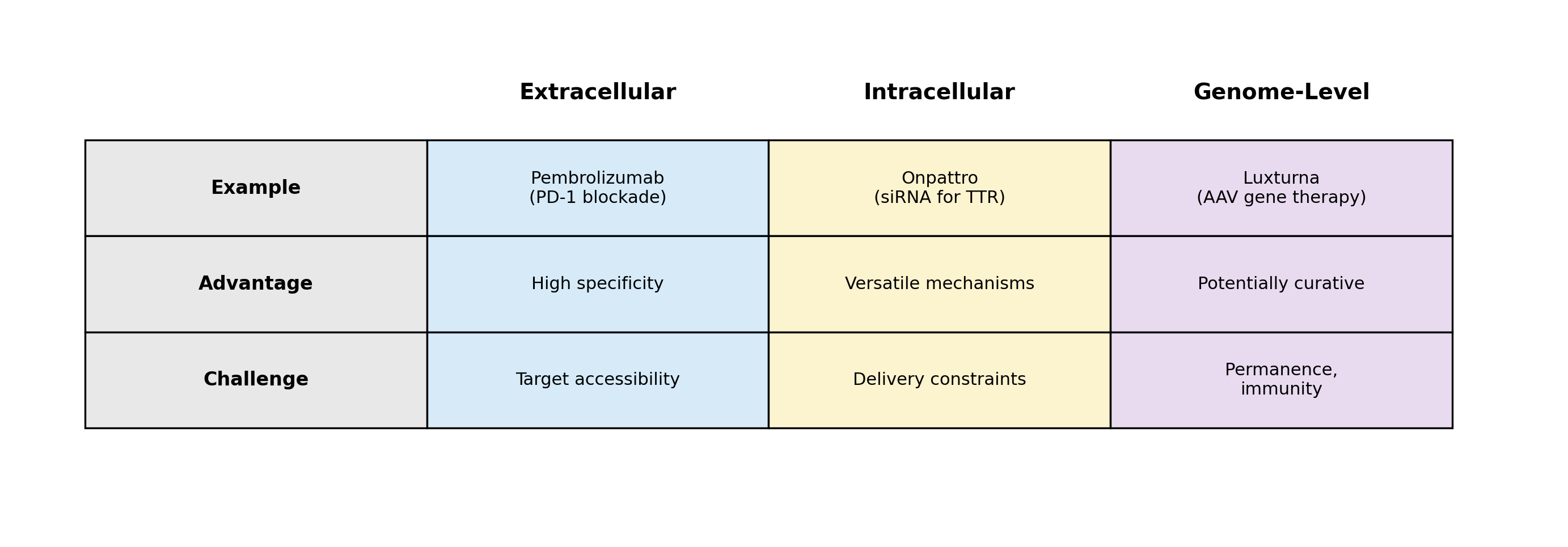
1. Extracellular Biologics
Examples: antibodies, bispecifics, cytokine therapies, receptor traps, CAR-T recognition domains
These biologics act outside the cell or on the cell surface, where targets are physically accessible and structure–function rules are relatively predictable.
Opportunities
- High specificity driven by well-understood protein–protein geometry
- Large-scale screening (phage, yeast, mammalian display)
- Modular engineering of binding regions
- Antibody formats (monoclonals, bispecifics, fragments) allow rapid iteration
Constraints
- Limited to surface or circulating targets
- Poor access to intracellular biology
- Tissue penetration shaped by antibody pharmacology
- CAR-T introduces complex in vivo behaviour (expansion, persistence)
Extracellular biologics excel when the disease presents an accessible handle. When it does not, the modality cannot reach it.
2. Intracellular Mechanism Biologics
Examples: siRNA, antisense oligonucleotides, enzyme-replacement therapies, intracellular protein modulators
These therapeutics act inside the cell, altering transcription, translation, or biochemical pathways. Unlike extracellular biologics, they rely on mechanistic clarity, not discovery screening.
Opportunities
- Can modulate almost any intracellular process
- Sequence defines function, enabling rapid design cycles
- siRNA therapies offer precise gene silencing
Constraints
- Delivery dominates feasibility (LNP tropism, endosomal escape)
- Misunderstood mechanisms scale into systemic toxicity
- Effects are transient unless re-dosed
- Enzyme therapies must navigate stability and trafficking challenges
Here, the central question is what mechanism the cell should execute. Once that is defined, the remaining challenge is delivering the payload to the right cell and compartment.
3. Genome-Level Biologics
Examples: AAV and lentiviral gene therapies (viral vectors for delivering DNA), CRISPR editing, base and prime editors, engineered stem cells
These interventions modify DNA itself or deliver genes for long-term expression. They operate at the deepest layer of biological control.
Opportunities
- Potentially curative, one-time interventions
- Ability to replace, silence, repair, or reprogram genes
- Ex vivo approaches (e.g., CRISPR-edited hematopoietic stem cells) enable precise engineering
Constraints
- Delivery is tissue-specific and immunogenic
- Effects are long-lived or permanent
- Redosing is limited by vector immunity
- Off-target editing and manufacturing add complexity
Genome-level biologics do not adjust cellular state—they redefine the functional capacity of a tissue.
These layers are not static; innovation happens at their boundaries.
What Comes Next: The Expanding Edge of Living Drugs
New modalities are emerging at the boundaries of these layers.
Antibody–drug conjugates (ADCs)
Bring extracellular precision together with intracellular effect.
Lysosomal-targeting degraders (LYTACs and related systems)
Extend extracellular biologics into controlled degradation of surface proteins.
Programmable RNA medicines
mRNA and antisense oligonucleotides use RNA as an instruction layer, enabling sequence-directed expression or modulation. These transient interventions sit within the intracellular layer but reflect a mindset closer to genome-level design.
Biologics are shifting from static molecules to orchestrated systems — designed to bind, enter, edit, or signal.
Two Different Games: Chemistry vs Biologics
It is tempting to treat drug discovery as one optimisation problem. It is not.
Small molecules operate in a space of exploration: vast chemical possibilities, incomplete mechanisms, complex ADME behaviour, and nonlinear structure–activity relationships.
Biologics operate in structured spaces shaped by evolution. Their specificity is often intrinsic: structure, sequence, and biological constraints guide how they bind and what they affect. These modalities also inhabit defined architectures — antibodies, RNAs, and viral vectors are not expansive chemical spaces but engineered variants of biological scaffolds. Yet they share a common limitation: delivery. Whether through tissue penetration, cellular uptake, or vector tropism, getting a biologic to the right place often determines what is feasible at all.
Most importantly, extracellular and genome-level biologics can do things chemistry cannot do at all — surface modulation, gene expression, gene repair, engineered cellular behaviour.
The mathematics may converge, but the design problems diverge:
- different priors
- different constraints
- different observables
- different uncertainties
- different consequences of error
Thinking clearly requires understanding which game you are in — and which rules apply.